Quick Search
The search engine should enable you to search and browse the TRANSPATH database. By default, it will also query NetPro™ data (if licensed).This opportunity can be switched off. The basic search and retrieve function is a very easy one. First, choose the table you are interested in using the menu on the left. This will open the main search form.
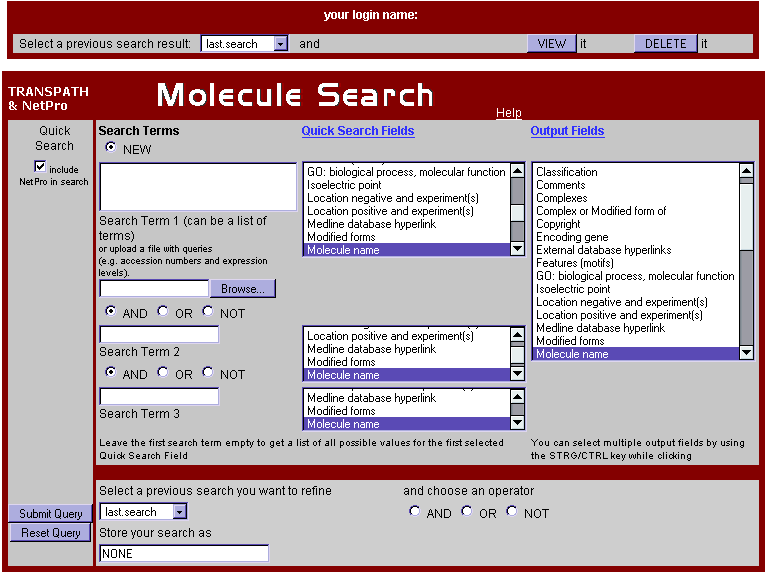
Fig.1 Molecule Search Form
Enter the terms with which you want to query TRANSPATH in the input boxes on the left. Here you can connect up to three search terms with the logical operators AND, OR, or NOT. Wildcards are set automatically in front of and behind every search term. For searching with lists of search terms (e.g. gene lists from microarray experiments) please fill into the top input box on the left and use a new line for every search term you have. You can also upload a text file with your search items.
Now choose the appropriate search fields in the lists in the middle, called Quick Search Fields. These lists show all field names that occur on the data sheets for the corresponding table.
You can specify the columns you want to appear in the result list by selecting fields in the list box (Output fields) on the right. Choose multiple output fields by holding the STRG/CTRL key while clicking. Press the help button below the list field to find out more about the meaning of each field name. After you have made your choice, click on the Submit Query button and the query will start.