MAIN MENU
The signaling network visualization tool for TRANSPATH®, the PathwayBuilder™, provides two different interfaces for creating specific views of the data. The Main Menu is a general interface that allows access to the functions of the program in an uncomplicated manner. The Expert Menu enables the user to select parameter settings manually and influence the visualization results in detail.
Scope
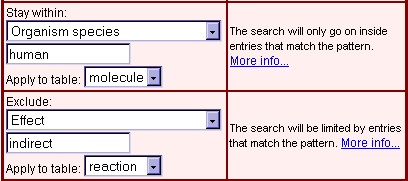
Selecting the scope of visualization with the Main Menu offers two main methods of application (Fig. 1). There is either the opportunity (show neighbours) to search and view all nodes (molecules, genes, and reactions) within an adjustable radius around the starting signaling component, or queries can be started for shortest paths to definable target nodes, which can be important biological groups (such as receptors, transcription factors, or genes), saved results of queries (e.g. microarray data sets) with the TRANSPATH search engine, or matches for free-text search terms.
|
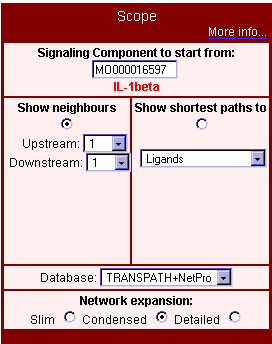
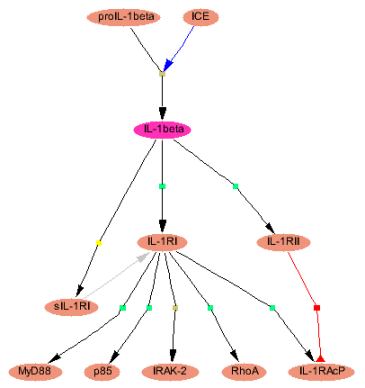
Fig. 1: Selecting the scope of visualization with the main menu of the PathwayBuilder™ interface
|
With the upstream and downstream parameters in the show neighbours section, the radius of the visualized neighbourhood regarding direction and distance, i.e. the number of steps upstream and/or downstream of the starting node, can be determined. One step is defined as the distance from one component to the next connected component of the same type (molecule or reaction). For example, if you start from a reaction downstream with a distance setting '1', the signal flow map will show you the starting point reaction, the signal acceptor molecule(s)/gene(s), and reactions extending from this molecule(s)/gene(s). Indirect reactions will consume 3 steps, this means to see indirect reactions going out from a molecule you have to set the downstream distance to at least '3'.
|
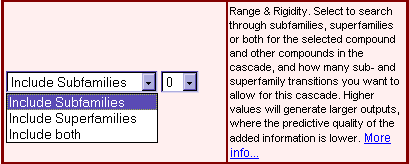
| The shortest path function is implemented to find the most direct connections to defined groups of molecules/genes. For example, to find the shortest path between IL-1beta (interleukin 1 beta) and IKK (inhibitor of kappaB kinase) molecules, select 'customized', 'name, synonyms' as field, 'molecule' as table and enter 'IKK' as target. If the target is outside of the standard radius settings (3 steps 'up', 3 steps 'down'), you can manually increase the search distance directly in the map (Fig. 4). Detected targets are depicted in blue-grey (Fig. 2: Shortest path from IL-beta to IKK molecules). To find the shortest connections to prominent functional groups of components such as transcription factors, simply select the group name (Fig. 3: Shortest paths from IL-1beta to transcription factors).
|
Table 1: The Expert Menu parameter settings that are used as standard settings in the Main Menu's network expansion levels 'Slim', 'Condensed', and 'Detailed'
|
| Main Menu
|
| Expert Menu parameters |
Slim |
Condensed |
Detailed |
| Include subfamilies |
0 |
1 |
1 |
| Show unmodified form |
off |
off |
on |
| Show bordering entries |
off |
off |
on |
| Condense basic molecules |
off |
on |
off |
|
The network expansion parameter with its three levels 'slim', 'condensed', and 'detailed' influences the number of connections that are shown. 'Slim' shows a network that stays on the hierarchical level of the starting component (Fig. 4) – reactions connected to superfamily or subfamily molecules are not taken into account. This selection is best for creating an overview. 'Condensed' allows the viewing of all reactions linked to basic molecules subsumed to their corresponding orthogroup entries. This option is most useful when starting with an orthogroup molecule. 'Detailed' shows a complex network including reactions linked to entries one hierarchical family level below and also follows the modified form links in reverse direction. Table 1 shows the corresponding parameter settings in the expert menu for the three levels of network expansion.
|