Search in one table (customizable search)
The search engine should enable you to search and browse the TRANSPATH database. By default, it will also query NetPro™ data (if licensed). This option can be switched off. Choose the table you are interested in, using the menu on the left. This will open the respective search form.
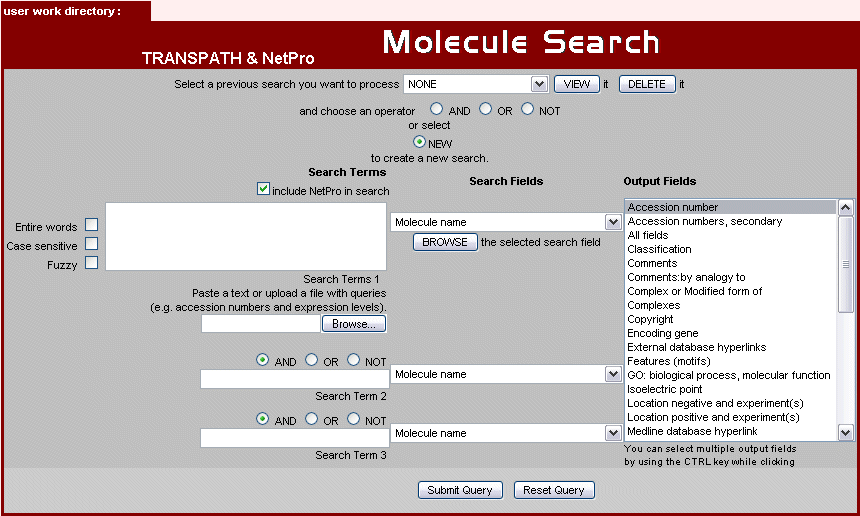
Fig.1 Molecule Search Form
Enter the terms with which you want to query TRANSPATH in the input boxes on the left. The Search Field, in which you want to search, can be selected to the right of the input fields.
For a description of the various fields, please see the description for the respective table in the documentation.
(The following fields are always conjointly searched:
in the MOLECULE table, the "molecule name" field together with the "synonyms" field;
and in the GENE table, the "gene name" field together with the "synonyms" field).
You can specify the columns you want to appear in the results list by selecting fields in the list box (Output Fields) on the right.
Choose multiple output fields by holding the CTRL (STRG) key while clicking.
After you have made your choice, please use the Submit Query button to start your search.