BIOBASE GmbH
Halchtersche Strasse 33
D-38304 Wolfenbuettel, Germany
| 1. For all UniGene clusters (G - is the total number of clusters for human genes), we collected all of the information about libraries that was used for sequencing the ESTs included in the clusters. | ||
| 2. From the list of libraries we excluded all cancer-related libraries. Only the libraries of normal tissues and organs were considered. | ||
| 3. All tissue and organ names used in the selected list of libraries were linked to corresponding terms in the Cytomer® database [1]. All libraries linked to the same term were grouped into "organ library groups" (e.g. liver library group). P- is the total number of formed organ library groups. | ||
| 4. Compute nij which is the number of ESTs of a gene i (Unigene cluster) linked to a organ library group j . | ||
| 5. Compute | 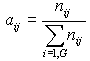 |
- an "abundancy score" of each gene in each group. |
| 6. Compute | 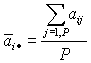 |
- an average "abundancy score" of gene i. And |
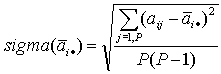 |
- standard error. | |
| 7. Compute | 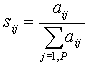 |
- a "specificity score" of each gene in each group. |
| 8. Compute | 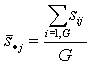 |
- an average "specificity score" of group j. And |
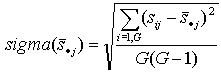 |
- standard error. | |
| 9. Compute | 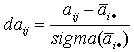 |
- an "abundancy parameter" which shows a difference of the abundancy score from an average abundancy score. In the case of daij ≥ 2.0, we can consider this difference as statistically significant. |
| 10. Compute | 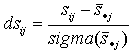 |
- a "specificity parameter" which shows a difference of the specificity score from an average specificity score. In the case of dsij ≥ 2.0, we can consider this difference as statistically significant. |
| 11. We used the following default cut-offs to establish links between genes i and tissue/organs j and to form groups of genes expressed in a given tissue/organ: daij ≥ 2.0 AND dsij ≥ 2.0 AND nij ≥ 2 . | ||
Wingender, E.
TRANSFAC®, TRANSPATH® and CYTOMER® as starting points for an ontology of regulatory networks
In Silico Biol. 4, 55-61 (2004).