|
|
 |
 |
|
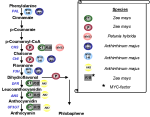
Figure 1: Conserved AA features of
Myb factors in plants.
The DNA binding domain consists of three 50 aa repeats in animals (R1-R3), whereas in plants only two repeats (in some cases only one) were detected. These repeats show most striking homology
to the animal R2 and R3. One repeat consists of 3 -helices (bars). The third helix (brown bar) of the R2 and R3 repeat is in contact to the major groove of the DNA molecule. Within these repeats a characteristic feature
is common to all MYB factors: the tryptophan cluster. Three tryptophan residues are spaced by 18-21 aa which are very conserved among all MYB factors (red boxes). |
|
|
 |
 |
|
Figure 2: Schematic presentation of
the most important steps in the synthesis of the pigments anthocyanin and phlobaphene. MYB factors of different species interact with promoter elements of several key genes (blue letters). The recognition sites of these
factors within the promoters are very similar. C1 activates his target genes only in presence of cell specific MYC factors (bHLH transcription factor).
Abbreviations: PAL Phenylalanine lyase, CHS
Chalcone synthase, CHI Chalcone isomerase, F3H Flavanone-3-hydroxylase, DFR Dihydroflavonol reductase, ANS Anthocyanidinsynthase, UF3GT UDP-Glycosyl-transferase |
|