 |

Introduction |
 |
 |
Fungi are not destructive and pathogens only, but also artists of surviving and can use a surprisingly diverse array of substrates such as nitrogen sources (ammonia, glutamine, glutamate, nitrate, nitrit, purines, amides, most amino acids, and proteins). Upon demand fungi are capable of expressing catabolic enzymes of many different pathways of primary and secondary metabolism.
Nitrogen control involves activation of the structural genes, which is prevented in the presence of prefered nitrogen sources. De novo synthesis of enzymes in a particular catabolic pathway is controlled at the level of transcription. Two zinc finger domains are a common motif for specific DNA-binding activities in fungal transcription factors. The zinc finger class comprises three groups: Cys2-His2 zinc fingers, Cys2-Cys2 zinc fingers, and binuclear Cys6 zinc finger.
In TRANSFAC database the amount of data of fungal metabolic regulators is permanently increasing. The entries contain detailed molecular and genetic information about protein structur, function, binding sites of the transcription factors. This is of interest for pharmaceutical industry, agriculture and medical research.
|

Cys2-Cys2 type |
| |
The DNA-binding domain of Cys2-Cys2 zinc finger proteins (Figure 1. Glucocorticoid receptor) is composed of two irregular antiparallel beta-sheets and an alpha-helix, followed by an extended loop. The zinc ion is coordinated by four cysteines in a tetrahedral manner. The proteins interact with DNA-elements containing the consensus target sequence A/T GATA A/G and were referred as GATA-factors. The helix and loop connecting the two beta-sheets bind in the major groove of the target DNA. Fungal GATA factors typically contain a single zinc finger (example of two zinc finger protein SREP in Penicillium) (Teakle & Gilmartin, 1998.)
|
|
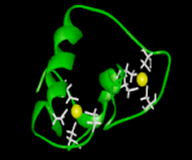
Fig. 1 Glucocorticoid receptor
(T. Sumiyoshi:www.Zf.htm) |
Examples:
TRANSFAC
NO. |
FACTOR |
ORGANISM |
BINDING SITES |
REG. FUNCTION |
| T025533 |
AREA |
A. nidulans |
|
+ Nitrogen |
| T00627 |
NIT-2 |
N. crassa |
24 |
+ Nitrogen |
| T02818 |
GLN-3 |
S. cerevisiae |
1 |
+ Nitrogen |
| T02411 |
DAL-80 |
S. cerevisiae |
3 |
- Nitrogen |
| T02406 |
URBS-1 |
U. maydis |
2 |
- Siderophore |
| T02819 |
WC-1 |
N. crassa |
|
+ Light |
Tab 1. Positive-acting global regulatory genes i.e. AREA in Aspergillus, NIT-2 in Neurospora, GLN-3 in Saccharomyces specify GATA-type zinc finger transcription factors which activate nitrogen structural genes. Some family members mediate responses to other signals, including light (Marzluf, 1997). |
|
|

Cys6 type |
| |
The DNA-binding domain of Cys6 proteins was first characterized in S. cerevisiae GAL-4 protein. It consists of six conserved cysteines, which form two alpha-helical structures separated by a proline-associated loop and it coordinates two zinc ions to form a cloverleaf shaped structure (Binuclear cluster) (Pan and Coleman, 1990). The DNA-binding sites consist of conserved trinucleotides, usually in a symmetrical configuration, they are spaced by an internal variable sequence of defined length e.g. GAL4 and LAC9 bind to CGGN11CCG, PPR1 (Fig.2) and UAY bind to CGGN6CCG (Todd & Andrianopoulos, 1997).
| C-X2-C-X6-C-X6-C-X2-C-X6-C |
|
|
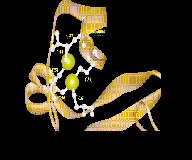
Fig.2. Subunit of PPR1
(T. Sumiyoshi: www.Zf.htm) |
Examples:
TRANSFAC
NO. |
FACTOR |
ORGANISM |
BINDING SITES |
REG. FUNCTION |
| T00302 |
GAL-4 |
S. cerevisiae |
30 |
Galaktose metabolism |
| T02841 |
FACB |
A. nidulans |
17 |
Acetate utilization |
| T02844 |
NIRA |
A. nidulans |
|
Nitrogen metabolism |
| T02845 |
NIT-4 |
N. crassa |
3 |
Nitrogen metabolism |
| T01163 |
PUT-3 |
S. cerevisiae |
1 |
Proline Utilization |
| T00346 |
HAP |
S. cerevisiae |
6 |
Oxygen metabolism |
Tab. 2. These Cys6 proteins are involved in a wide range of different processes of primary and secondary metabolism, like NIT-4 in Neurospora (nitrogen metabolism) and FACB in Aspergillus (acetate induction of acetamidase, acetate utilization enzymes acetyl-CoA synthase, activates isocitrate lyase and malate synthase) (Todd et al., 1998). |
|
|

Cys2His2-type |
| |
The Cys2-His2 proteins contain the zinc finger motif of TFIIIA/ Krueppel type. This predominantly eucaryotic DNA-binding motif was first discovered in the Xenopus transcription factor (TFIIIA) by Aaron Klug (1985). Therein two cysteines and two histidine residues coordinate one zinc ion. Each zinc finger module only comprises 30 aa and folds into beta-beta-alpha-motif, stabilized by chelation of zinc between a pair of cysteines by the beta-sheet and a pair of histidines by the alpha-helix. The alpha-helix binds to the DNA through the major groove.
| C-X2,4-C-X3-F-X5- L-X2-H-X3,4-H |
|
|
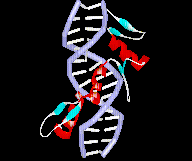
Fig. 3. Xenopus transcription
factor TFIIIA (T. Sumiyoshi:
www.Zf.htm) |
Examples:
TRANSFAC
NO. |
FACTOR |
ORGANISM |
BINDING SITES |
REG. FUNCTION |
| T00509 |
MIG-1 |
S. cerevisiae |
29 |
Glucose repression |
| T00011 |
ADR-1 |
S. cerevisiae |
21 |
Activator of perox. genes |
| T016679 |
PACC |
A. nidulans |
6 |
Act. of alkaline-exp. genes |
| T01258 |
MSN-4 |
S. cerevisiae |
3 |
Carbon utilization |
| T00776 |
SWI-5 |
S. cerevisiae |
2 |
Cell cycle regulator |
| T00731 |
RME |
S. cerevisiae |
1 |
Repressor of meiotic genes |
Tab. 3. A large number of developmental regulators are involved in cell cycle control. In Saccharomyces the zinc finger proteins control metabolic capabilities or regulation of cellular development (control of mating type). ADR-1 in Aspergillus activates peroxisomal protein genes. |
|
|
 |

Status of TRANSFAC data base |
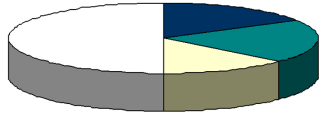
| At the moment the TRANSFAC database comprises 101 main entries of fungal regulators involved in important reactions of primary and secondary metabolism. Furthermore 230 binding sites are linked to these main factors. Different fungal transcription factor classes, including the zinc finger proteins, are inserted, like leucine zipper factors (bzip, zip), homeobox proteins, tryptophan cluster (Figure 4.). Currently we are working on the completion of the data. |
Fig.4. Fungal metabolic regulators in TRANSFAC data base (March 2000)
 |

April 2000 |
|






